Molecular Docking Software Design from a Computer Programmer’s Perspective
- mansour ansari

- Aug 19, 2025
- 2 min read

Molecular Docking Software Design from a Computer Programmer’s Perspective
It took me 7-8 months to build a working drug discovery system using my proprietary quantum-enhanced pipeline. The Quantum Pipeline, however, took about a year of my life, but it was worth it. The system I created is attached to my quantum entropy,
The work begins with the crucial stage of identifying potential drugs. In this short post, I try to explain how to build a similar system using classical RNG. For the Quantum integration, I am ready to help you build a scaffolding of a quantum interface right into your Drug Discovery system. That part is not covered in this short post.
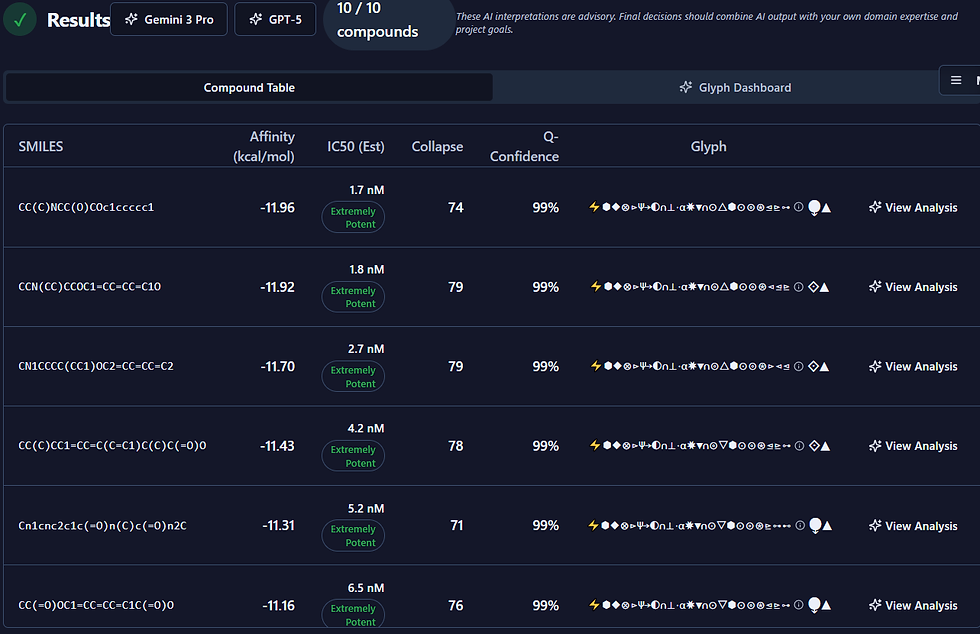
So, when people think about molecular docking, they picture biochemists in lab coats. From my angle, it feels more like software architecture with a strong physics engine under the hood. Docking is just code, compute, and clever planning. In fact, I built my system using a few old computers. Nothing fancy.

What You Need to Start
Programming skills (Python, C++, containerization). My skills are not in C++, Python, however is my thing. I also use the React 18 Engine.
Chemical toolkits (RDKit, OpenBabel, AutoDock/Vina binaries)
Protein/ligand data sources (PDB, PubChem, ChEMBL)
Compute resources (CPU clusters or cloud VMs, GPUs if ML is involved)
Entropy source (PRNG/QRNG for stochastic exploration). Ask me about the QRNG integration.

How to Plan for AI Submission
Data Preparation: Generate consistent ligand representations (SMILES → 3D conformers).
Feature Extraction: Transform docking results into descriptors AI models can digest.
Task Design: Define clear ML objectives (rescore poses, predict binding affinity, or classify novelty).
Audit & Provenance: Track seeds, versions, and metadata for reproducibility.
Pipeline Integration: Containerize so docking outputs feed seamlessly into AI jobs (local or cloud).

In short: think like a systems architect. Docking software is just another distributed application—except the end goal isn’t rendering pixels or serving ads; it’s bending molecules toward curing disease.
Next, I will include a Pseudocode version of a Drug Simulation architecture. I used a similar design in my system.


Comments