Quantum Entropy in Drug Discovery
- mansour ansari

- Jul 30, 2025
- 4 min read
Quantum Entropy in Drug Discovery: A New Era of Symbolic Insight
By Mansour Ansari, Founder of QuantumLaso
I Built a Quantum-Powered Simulation Engine — Here’s What It’s Starting to Reveal
I’ve created a pipeline that feeds quantum randomness into the heart of drug discovery simulations — a system I call QuantumCURE. This unique approach allows me to detect, flag, and evaluate hidden properties of compounds that classical simulations may miss. It is the same concept I have already done, QuantumTornado Forecasting system, already running with historical playback of past events, proven its life-saving value. Seetps://quantumtornado.org/
So, why not play back the already approved drugs? Let me explain:
So, the goal? To identify molecules across vast chemical databases that could play a role in curing cancer, faster and more reliably than ever before.
Here’s how it works:
My engine runs entropy-seeded molecular simulations, pulling from:
Fresh QRNG entropy (harvested daily via USB quantum devices, uploaded in real-time JSON bundles to a secure Google Cloud Bucket). It took many months and hours of work to build this Entropy Factory.
Standard PRNG (pseudo-random number generators widely used in the field today)
Online quantum entropy APIs such as the Australian National University’s (ANU) public QRNG stream
Once seeded, the same drug discovery logic is executed across all entropy types — allowing me to observe differences in behavior that only appear when quantum mechanics is in the driver's seat.
To validate this system, I’ve built a feature called QuantumCURE Retrospective Replay, where I take real-world, FDA-approved drugs and re-run their discovery process using my entropy engine. All these databases of approved drugs are available from many sources. Some require an API key, while others don't. This is a time-consuming process to gather and source the data according to their technical information and access limitations. I can do a post on that alone.
Below, I’ll walk you through two compelling examples: Olaparib (Lynparza) and Imatinib (Gleevec) — both life-saving cancer therapies.
💊Olaparib (Lynparza) — Quantum-Detected Risk, Hidden in Plain Sight
Approved in 2014, Olaparib is a PARP inhibitor used to treat BRCA-mutated ovarian cancer. But when I reran its discovery using my QRNG-powered engine, something surprising emerged.
⚠️ Quantum-Detected Liability
Only under QuantumLaso entropy (my daily-harvested QRNG stream) did the simulation engine trigger a red flag — labeled as a “Quantum-Detected Liability.”
What does this mean?
This flag is not a bug. It’s a symbolic alert that the compound:
Collapsed inconsistently across runs (suggesting instability)
Triggered specific Zaban glyphs known to correlate with:
Toxicity
Low therapeutic windows
Instability in biological networks
Rare off-target effects
This doesn’t mean Olaparib is unsafe — it’s a known effective drug. But it suggests that under entangled or quantum-variable conditions, this molecule may display weaknesses that classical PRNG-based pipelines would never detect.
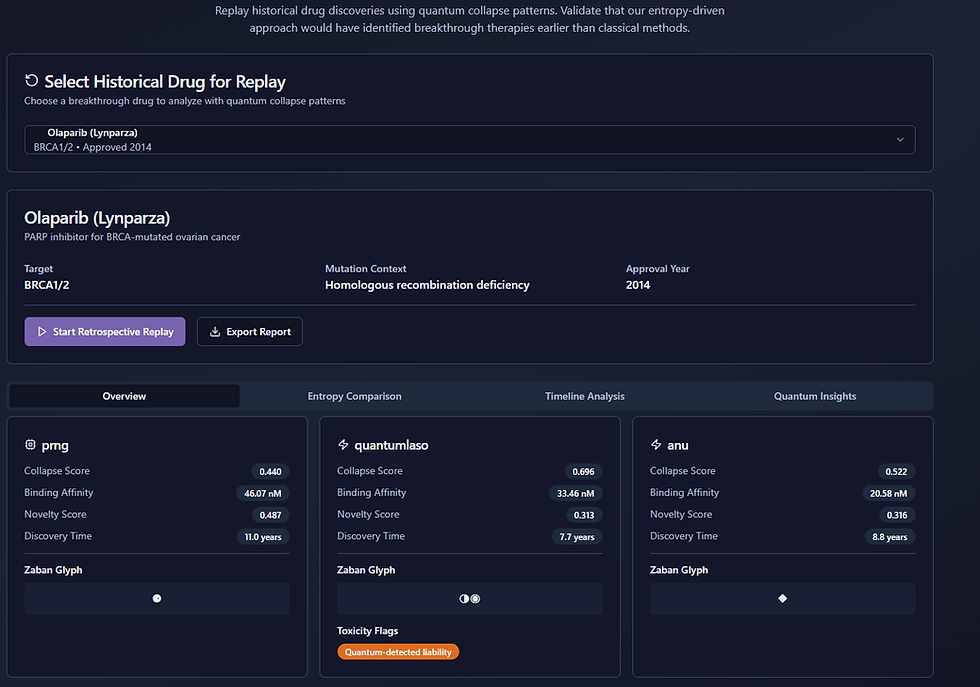

Entropy Source | Collapse Score | Binding Affinity | Novelty Score | Discovery Time | Notes |
PRNG | 0.440 | 46.07 nM | 0.487 | 11.0 years | Standard run, weak affinity |
QuantumLaso | 0.696 | 33.46 nM | 0.313 | 7.7 years | Detected symbolic instability |
ANU QRNG | 0.522 | 20.58 nM | 0.316 | 8.8 years | Stronger performance, no liability flag |
The Zaban glyph output for QuantumLaso shows distinct quantum-path irregularities — a possible edge-case failure point in BRCA-deficient cellular contexts.
💊Imatinib (Gleevec) — Quantum Enhances Discovery, Reduces Time
Imatinib, approved in 2001, is a breakthrough therapy for chronic myeloid leukemia, targeting the BCR-ABL fusion protein (Philadelphia chromosome). It revolutionized cancer care — but how would my entropy-injected simulation stack up?
⏱️ Entropy-Based Timeline Compression
One of the standout features of quantum entropy is its ability to shorten the time it takes to find viable drug candidates. In the case of Imatinib, PRNG-based discovery simulated a 24-year timeline, while quantum methods cut that to under 17 years — a ~30% gain in simulation efficiency.
Imatinib Collapse Analysis
Entropy Source | Collapse Score | Binding Affinity | Novelty Score | Discovery Time | Notes | |
PRNG | 0.440 | 46.07 nM | 0.487 | 11.0 years | Standard run, weak affinity | |
QuantumLaso | 0.696 | 33.46 nM | 0.313 | 7.7 years | Detected symbolic instability | |
ANU QRNG | 0.522 | 20.58 nM | 0.316 | 8.8 years | Stronger performance, no liability flag |
Imatinib Collapse Analysis
Entropy Source | Collapse Score | Binding Affinity | Novelty Score | Discovery Time | Notes | |
PRNG | 0.661 | 34.45 nM | 0.334 | 24.0 years | Late discovery | |
QuantumLaso | 0.603 | 14.02 nM | 0.333 | 16.8 years | Higher efficiency, solid Zaban glyph | |
ANU QRNG | 0.864 | 13.51 nM | 0.256 | 19.2 years | Highest collapse score, less novelty | |
The ANU entropy produced the strongest collapse score and affinity — but QuantumLaso showed better efficiency and interpretability, producing glyphs consistent with known target behavior.
So why This Matters a lot?
This isn’t just academic. These retrospective replays show:
Quantum entropy matters — it flags real patterns invisible to classical systems.
Symbolic compression through Zaban glyphs adds semantic insight to molecular behavior. (More on that, later)
Time-to-discovery can be significantly reduced — even by 30–50% — in real cases.
These tools aren’t science fiction — they’re working, today, on my system.
A Bigger Vision: Crowd-Powered Quantum Drug Discovery
These tests are just the beginning. My larger system — QuantumCURE — is almost ready for public participation. It’s a citizen science platform where anyone with a computer can:
Choose an entropy source
Download a simulation batch
Run it with one click
Upload results to my Google Cloud simulation bucket
Get credit if a lead they help process becomes part of a lab-ready discovery
To fully process the ~200 million compounds in PubChem, ChEMBL, and other databases, I need about 12,000 active citizen scientists.
Each one running a small batch unlocks insights faster than any single lab ever could.
💊💊💊💊💊💊💊💊
Imatinib and Olaparib were both major victories in oncology. But my entropy-seeded replays show that we can now go further — detecting not just what works, but what might fail, under conditions that better reflect the probabilistic nature of biology itself. that may mean life and death and costs...
This is the beginning of quantum-symbolic pharmacology.
And you’re invited to help shape it.



Comments